TQL Functional Description
Introduction
TQL is is designed as a language in which to formulate queries against terminologies, generating 'result subsets'. A TQL query does not say just what code points will be in the subset, but what the structure of the subset will be. This structure is determined by a combination of relationships from the terminology (such as IS-A) and structuring relationships defined in the query. A TQL result subset is in general, like a small version of its source terminology, containing semantic relationships intact. This allows the subset to viewed and traversed and therefore used for choosing terms, in a similar way to its parent terminology.
The facilities of TQL can be understood by a set of examples, which are here taken using the SNOMED-CT terminology from IHTSDO.
Basic Concepts
PassThrough, Navigate, Exclusion
In the following example, a TQL query is used to generate a subset of blood types from SNOMED.
The design intention of the query is as follows:
- Ease of viewing and navigation to common blood type categories;
- Removal of unwanted 'Kidd' blood type categories;
- Grouping of all minor / unusual blood types in a separate tree.
- Otherwise retain SNOMED-CTs subsumption structure.
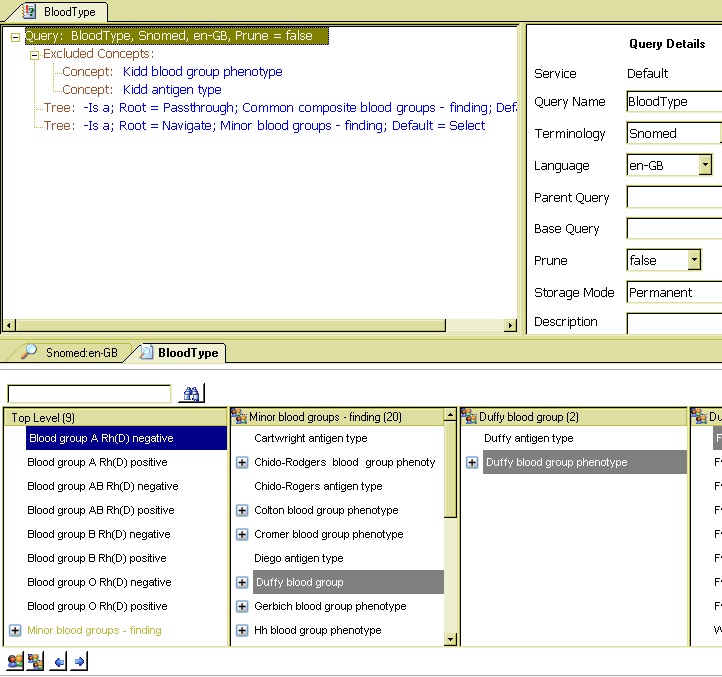
The following screenshot shows the query definition and also the result in a viewer at the bottom. These requirements are satisfied as follows. The query is defined as two 'trees', the first representing the common groups. For ease of viewing and navigation, the root node of this tree ([365640002 | Common composite blood groups - finding]) is set to passthrough, meaning 'pass through this node when generating the subset'. The effect is to show the children of the tree directly, exactly as required for fast access by clinical users.
The second tree has as its root node the [365640002 | minor blood groups] node from SNOMED. In this case, the root node is set to navigate, meaning 'retain it when generating the subset, but only for navigation purposes, not choosing purposes'. It is accordingly shown in a light colour in the viewer and is not selectable.
Lastly, two exclusions are made, of the categories [115830003|Kidd blood group phenotype]. The view at the bottom shows that the remaining requirement has been met - the hierarchy is the SNOMED subsumption structure for the minor blood group types.
Descriptive Nodes, Iterators, Related Concepts
In the following example, a TQL query is defined to generate the result subset corresponding to 'all bacteria'. The design intention here was as follows:
- Given the very large number of matching concepts in SNOMED, provide hierarchical navigation by 2 top-level pseudo-categories (i.e. that don't exist in SNOMED), namely 'By phylum' and 'Common categories', replacing the SNOMED node 'Bacteria', which is then of no practical use;
- For phyla relating commonly to human health, such as Chlamydiae, force selection of a particular sub-node at at least the 'Order' level (Chlamidiales)
The query definition shown in the screenshot below illustrates the use of several query node types in defining the two pseudo-categories. Both start with a 'Descriptive Node'. These nodes will be inserted into the result subset but are marked for navigation only - they cannot be selected by an end user browsing the result subset for use as a terminology concept. The first pseudo-category define (By Phylum) is created through a structure which:
- Uses a 'Concept Node' to specify that the [41146007 | Bacteria] concept should be inserted into the result subset directly under the 'By phylum' descriptive node.
- Uses an 'Iterator' which essentially functions as a loop - all the nodes below the Iterator will be repeated in building the result subset as long as each iteration adds at least one concept to the result subset.
- Within the Iterator uses a 'Related Concepts' node to specify that each level of the hierarchy generated by the iterator is formed by following the 'IsA' relationship within the terminology concept hierarchy. The end result of the Iterator plus the Related Concepts node is to build a hierarchy in the result subset formed by following the chains of IsaA relationships in the terminology.
Although the Iterator node can be used to build very powerful queries there is a simpler way to accomplish building a simple hierarchy. This is the 'Tree' node. Its use is illustrated in defining the second pseudo-hierarchy (Common categories), whose root point is [409822003 | Bacteria].
Advanced Concepts for Terminology Management
Special Nodes in a Tree
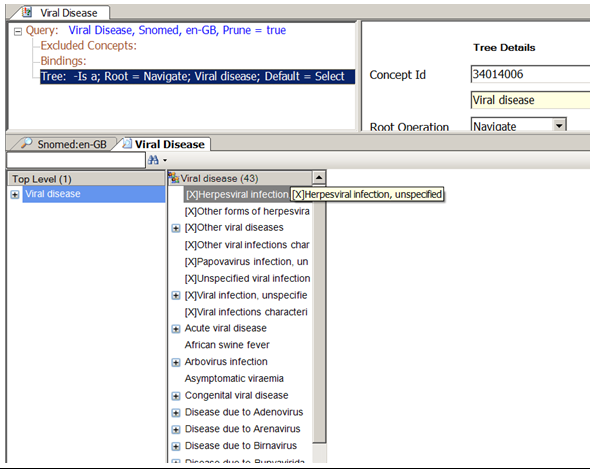
In the following example, a TQL query is defined to generate the result subset corresponding to the 'Viral Disease' query . The design intention here was to show the hierarchy of concepts below the 'Viral Disease' concept (concept 34014006). Two versions are shown in the screenshots below: the first is the complete hierarchy beneath the concept, the second shows the query without any of the 'Limited' SNOMED CT concepts (these concepts all have a concept status of 6).
The query specifies the hierarchy via a Tree node and uses the 'Special Node' to specify that concepts with a status of '6' should be excluded from the result subset.
Bindings
Multiple variants or terminology sources can be bound in one query, where a 'source' is a terminology (with stated version), another query, or a concept hierarchy within a terminology. Each binding is tagged with a symbolic name that can be used throughout the query to scope other statements.
Comparing two different releases of a terminology
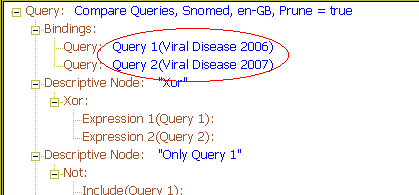
A common use of binding is to build queries that show differences and commonality between two given releases of a terminology. The following example shows how two releases of SNOMED-CT are used within the query - 31 Jan 2006 and 31 Jan 2007, bound respectively to the names "Release 1" and "Release 2". The goal of this query is to compare the two releases in the hierarchy [34014006 | Viral disease].
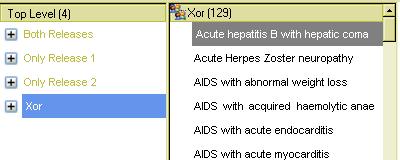
In the query, four top-level nodes are defined using various Boolean operators, each defining a subset of terms in the descent of Viral disease. The Xor node determines what terms are only in one release but not both; the Not operator is used twice to define terms only in one or other release respectively, and the And node finds terms common to both.
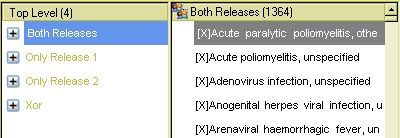
The results can be seen below. The first view shows the Xor node result, containing 129 terms that occur only in one or other of the releases in Viral Disease.
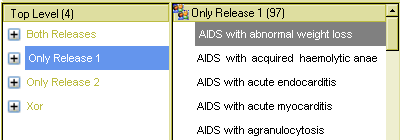
The next view shows 97 terms only in the older release, i.e. terms that have been obsoleted or otherwise removed in the newer release.
The following view shows the reverse, i.e. 32 terms new in the more recent release.
The last view shows the 1364 terms common to both releases, in the Viral disease hierarchy.
Note that within the tool, a list-flattening option was chosen, however, the query results could also have been left in their original tree structure.
Comparing Two Queries
Two previously defined queries can be compared using a third query, using the binding method, this time to each of the queries rather than source terminologies. In the following example, bindings are to two queries that select the same descent - Viral disease - as the example above, from the same two releases of Snomed. The query results are identical to those above.
Other operations on two queries
However, this method can be used to perform other logical operations on existing queries, such as adding their results (Or operator), finding results common to both (And) operator, or finding results from only one or other.
Another kind of comparison that can be made is between two versions of the same query, i.e. which are different due to development work over time. As long as each version is checkpointed (saved), it will be identified and become available for this purpose (see below).
Comparing Hierarchies within a Terminology
A third operation based on the use of bindings allows two hierarchies within a terminology to be compared. This is useful in multi-hierarchy terminologies such as SNOMED-CT, because it enables a terminology developer to find terms in both hierarchies, and only in one or the other. The following example shows a comparison between the two concept hierarchies [102957003 | Neurological finding] and [118240005 | Finding by method].