...
One of the challenges of ontological models is that to work, the data on which inferencing is to be done using the ontology must themselves have a meaning consistent with the ontology. In practical terms this means that the information model(s) of the data must be consistent with or mappable to the ontologies; it also means that the data themselves are likely to be tagged with terms from an ontology. For example, if the data record a 'allergy' for a patient, this must have the same meaning as 'allergy' does in the ontology. However, this is often not the case due to poorly defined terms; 'allergy' might have been used to mean 'an allergic reaction' or 'a diagnosed allergy. Ontologies can help here by allowing the detection of such ambiguities (see http://ontology.buffalo.edu/medo/Cologne.pdfandpdf) and by providing well-tested guidelines for how to deal with the corresponding distinctions
Ontologies also exist in software, although most software developers have no idea of this, due to the failure so far of mainstream ICT education to take account of semantics within technical models (i.e. 'class', 'object' or E-R models in the programming sense). Nevertheless, everytime any 'modeller' or programmer creates code, a UML model or an information schema, they are creating some kind of ontology, usually of informational concepts. Software models should be understood as ontologies, because they make commitments to particular notions of the concepts they model - for example kumho solus kr21 , the base data types (Integer, Boolean etc) of programming languages.
...
If you have never seen an ontology before, you may find John Sowa's top-level categories interesting - this is a well-known example of an upper ontology. Needless to say, this is regarded as by no means the best or most relevant in the biomedical sphere, but it is a useful reference point.
...
- 'ontologies of reality' - ontologies whose subject matter is real things, processes or events, rather than information
- 'ontologies of information' - ontologies whose subject matter is information of any kind - i.e. utterances committed to a medium. Concepts undelying underlying such an ontology are likely to have to do with the process of investigating, recording, reporting or similar ideas.
...
In openEHR we are interested in ontologies of both kinds. Since the EHR is about recorded information, ontologies of the second kind, of information, are relaventrelevant. However, within recorded information of course we expect to find:
...
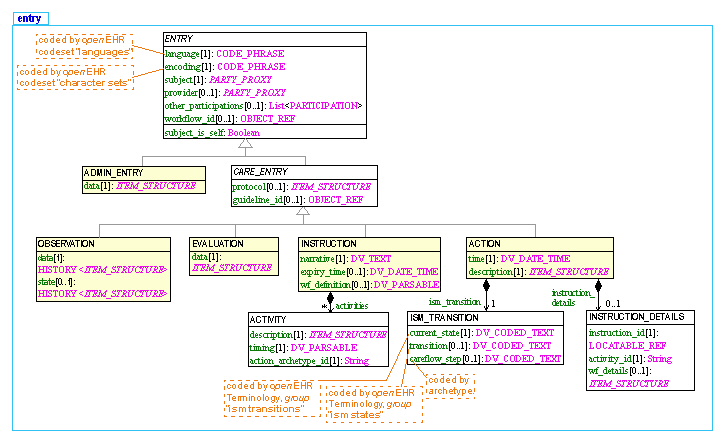
The openEHR Reference Model includes an information model which defines many classes. The information model, although relatively basic with respect to some of the ontologies in the biomedical space, nevertheless contains ontological commitments of its own - i.e. formal descriptions of certain real-world concepts relating to recorded health information. The part of the model of most ontological interest is the 'Entry' part, which is formally specified in the openEHR EHR Information Model; it can also be seen online as detailed UML. A summarised UML form of the Entry types in openEHR is illustrated below.
| There are 5 types of ENTRYs defined: an ADMIN_ENTRY type and 4 sorts of CARE_ENTRY types: OBSERVATION, EVALUATION, INSTRUCTION, and ACTION. The 6th type is called GENERIC_ENTRY, and is designed for mapping into and out of legacy and integration structures such as CEN EN13606, HL7 CDA, message and relational databases. The UML model of this type is here; it is documented in the openEHR Integration IM. All of these types are extremely generic and archetypes are used to define the specific business/domain content models under each of these types - see archetype mindmap for examples. |
The openEHR Entry model the way it is today has an ontological design which is described in a MedInfo 2007 paper. We can summarise the approach with the following three diagrams. The first is a model of process (shown in two ways); the second shows a cycle of information creation due to the process, and the last shows the information ontology developed in openEHR for recorded clinical information.
...
The explanation of the ontology behind the openEHR Entry model is in MedInfo 2007 paper. The following 3 diagrams show in a summarised way the ontological underpinning of the openEHR Entry model the way it is today. The first is a model of process, the second a model of information creation due to this process, and the third an ontology of information.
...
...
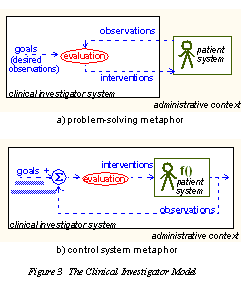
| We model health care delivery as two kinds of process: a clinical process , (corresponding to the interaction between a 'clinical investigator system' and a 'patient system', ) situated within a business process, which is owned by an 'administrative context'. The clinical process constitutes a sub-process of the business process, i.e. it is the main mechanism for the business process to achieve its goal, which is to satisfy a demand for care on the part of the patient. The administrative context corresponds to the health system as a whole, rather than a single enterprise, since from the patient care point of view, the mobilisation of care delivery is carried out by a network of provider organisations. The model can be illustrated in two equivalent ways, as shown in Figure 3. |
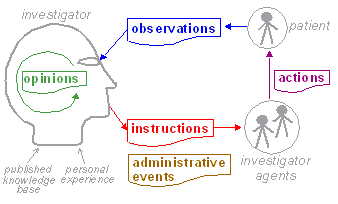
| The terms 'observation', 'evaluation' etc defined above are not themselves the same as information types, since they refer to a variety of phenomena within the process: information from observations, the activity of evaluation, acts of intervention, and goal statements. To be more precise, we are mainly interested in information created by the investigator system, since this notional agent encompasses any person or device who/which performs any healthcare related task, including the patient herself. The investigator system is therefore the creator of all clinical information in the health record, including patient-entered data. A small amount of administrative information may also end up in the EHR, generally created by non-clinical actors in the organisational context.
|
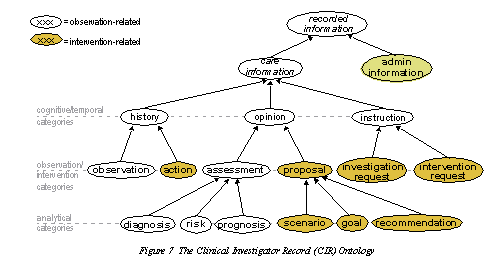
| The Clinical Investigator Record (CIR) ontology: |
...
In the openEHR approach, most description of the contents of recorded health information is left to archetypes (openEHR FAQ). An archetype can be thought of as a model of some clinical content (e.g. what is recorded in a urinalysis, or an ante-natal visit), expressed in a constraint formalism known as ADL (which has some similarities to OWL). Over 200 archetypes have been defined during NHS projects, Australian GP projects, and openEHR activities (openEHR archetypes page). To go straight to the point, an ontological way of looking at the archetypes that exist is the mindmap view. The structure of each archetype can be viewed by clicking on a node in this view. Another way to view archetypes is with the ADL workbench tool, and with various archetype editors. Example archetypes: Microbial lab observations; Adverse reaction; Examination of named body part.
...
The Archetypes and terminologies can be related directly in two ways (described in detail elsewhere):
- A node in an archetype which has a meaning represented in a terminology may be bound to that terminology and the relevant concept; and
- A node in an archetype may constrain the values of a DV_CODED_TEXT to be a specified set of terms from a particular terminology.
...
Rector A L, Nowlan W A, Kay S. Foundations for an Electronic Medical Record. Methods of Information in Medicine, 1991, 30:179-186. citeseerAnchor Rector1991 Rector1991
Austin JL. How to Do Things With Words. Cambridge (Mass.) 1962 - Paperback: Harvard University Press, 2nd edition, 2005. amazonAnchor Austin1962 Austin1962
HL7 International. Reference Information Model (RIM). See http://www.hl7.org.Anchor hl7org hl7org
Weed LL. Medical Records, Medical Education and Patient Care. The Problem Oriented Medical Record as a Basic Tool. Cleveland: Case Western Reserve University press, 1968. google scholar amazonAnchor weed1968Weed1968weed1968 Weed1968
Ceusters W, Smith B. Strategies for Referent Tracking in Electronic Health Records. J Biomed Inform. 2006 Jun;39(3):362-78. (ePub 2005 Sep 9, draft, slides presented during the IMIA WG6 workshop Ontology and Biomedical Informatics, Rome, Italy, April 29 - Mai 1, 2005)Anchor ceusters_smith_2006 ceusters_smith_2006
Rudnicki R, Ceusters W, Manzoor S, Smith B. What Particulars are Referred to in EHR Data? A Case Study in Integrating Referent Tracking into an Electronic Health Record Application. In Teich JM, Suermondt J, Hripcsak C. (eds.), American Medical Informatics Association 2007 Annual Symposium Proceedings, Biomedical and Health Informatics: From Foundations to Applications to Policy, Chicago IL, 2007;:630-634. (abstract, draft)Anchor rudnicki_et_al_2007 rudnicki_et_al_2007
Elstein AS, Shulman LS, Sprafka SA. Medical problem solving: an analysis of clinical reasoning. Cambridge, MA: Harvard University Press 1978. amazonAnchor Elstein1978 Elstein1978
Tange HJ, Dietz JLG, Hasman A, de Vries Robbé PF. A Generic Model of Clinical Practice - A Common View of Individual and Collaborative Care. Methods of Information in Medicine 3/2003. Schattauer GmbH. complete articleAnchor Tange2003 Tange2003
Bruun-Rasmussen M, Bernstein K, Vingtoft S, Nøhr C, Andersen SK. Quality labelling and certification of Electronic Health Record Systems. Studies in Health Technology and Informatics 2005; 116: p47-52. pubmedAnchor Bruun2005 Bruun2005
Areblad M, Fogelberg M, Karlsson D, Åhlfeldt H. SAMBA - Structured Architecture for Medical Business Activities. In: Engelbrecht R, et al. (editors) Connecting Medical Informatics and Bio-Informatics. MIE 2005: Proceedings of Medical Informatics Europe; 2005 Aug 28-31; Geneva, Switzerland. p. 1225-30.Anchor Areblad2005 Areblad2005
Beale T, Heard S. The GEHR Object Model - Technical Requirements. 2000. complete document.Anchor Beale2000 Beale2000
RICHE Consortium. RICHE ESPRIT Project. Final Report. Nov 30 1992.Anchor Riche1992 Riche1992
Ingram D, Lloyd D, Kalra D, Beale T, Heard S, Grubb, P, Dixon R, Camplin D, Ellis J, Maskens A. Deliverable 19,20,24: GEHR Architecture. GEHR Project 30/6/1995. complete document.Anchor Ingram1995 Ingram1995
Pierre GRENON, Barry SMITH and Louis GOLDBERG. Biodynamic Ontology: Applying BFO in the Biomedical Domain. From D. M. Pisanelli (ed.), Ontologies in Medicine, Amsterdam: IOS Press, 2004, 20-38. complete articleAnchor Grenon2004 Grenon2004
Thomas Beale, Sam Heard. An Ontology-based Model of Clinical Information. pp760-764 Proceedings MedInfo 2007, K. Kuhn et al. (Eds), IOS Publishing 2007.Anchor Beale2007 Beale2007